NO.201 Deep Tissue Mining: A Roadmap to Enable the Curation, Analysis, and Visualization of Petabyte-Scale Whole-Organ Multiplex 3D Tissue Images
June 2 - 6, 2024 (Check-in: June 1, 2024 )
Organizers
- David Mayerich
- University of Houston, USA
- Nancy Amato
- University of Illinois at Urbana-Champaign, USA
- Jinhyun (Jinny) Kim (김진현)
- Korea Institute of Science and Technology, South Korea
- Markus Hadwiger
- KAUST, Saudi Arabia
- Guoning Chen
- University of Houston, USA
Overview
Overview
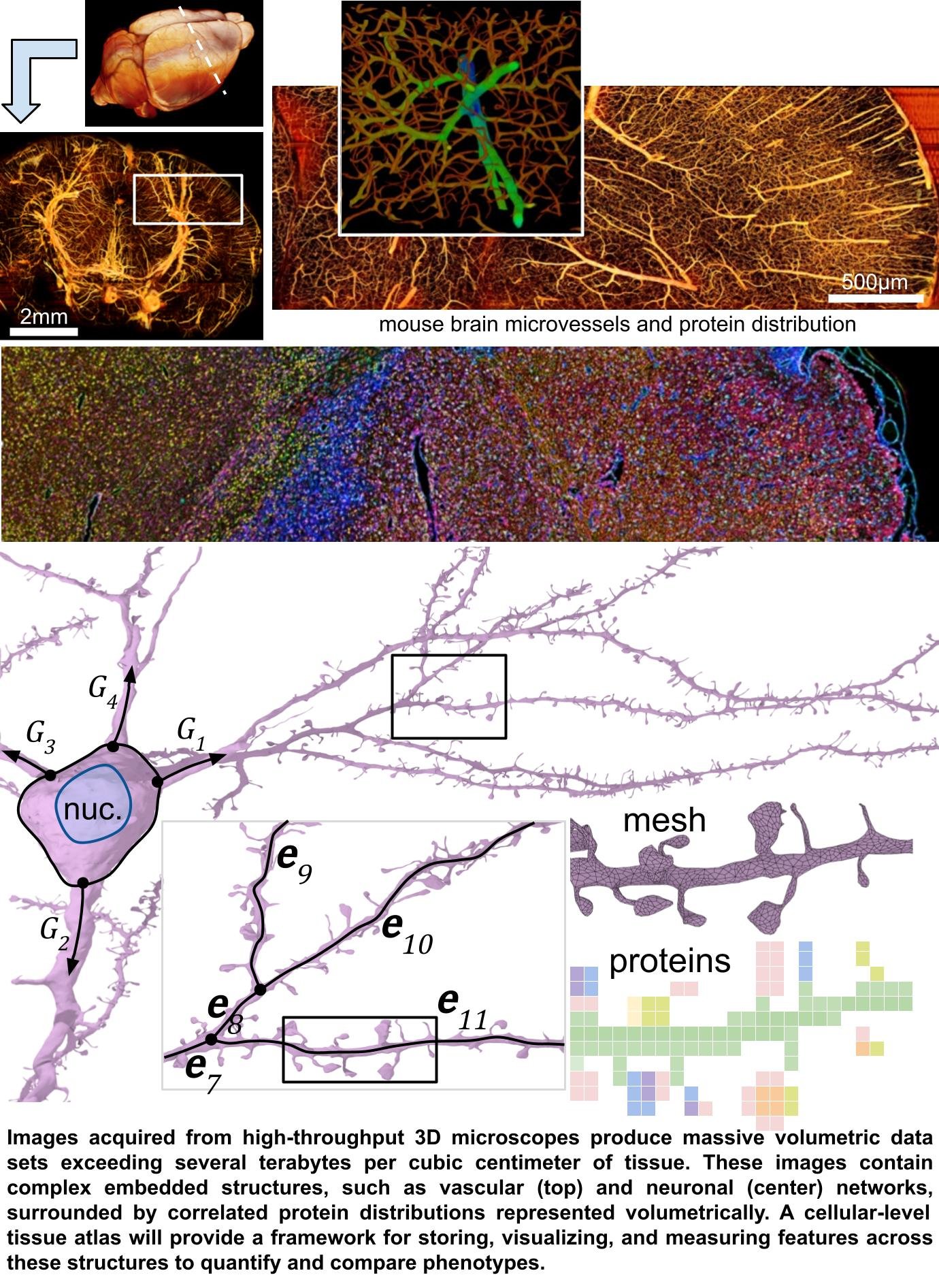
Whole-organ cellular-level tissue atlases will fundamentally impact biomedical research and education in the same way that satellite imagery and global positioning have changed navigation. Whole-organ tissue atlases will lead to breakthroughs in research and precision medicine, and an established community-endorsed software framework is critical to their construction. However, the community currently lacks the infrastructure to cope with petabyte-scale volumetric data with complex embedded structures - such as vascular and neural networks combined with sparse volumetric protein distributions. This meeting will formally initiate the development of a new field focused on whole-organ cellular-level mapping, including visualization, reconstruction, and analysis of massive whole-organ multiplex volumetric data at petabyte scales.
There is a compelling need to study biological systems at the whole-organ scale with the benefit of spatially-distributed deep molecular signatures representing hundreds to thousands of proteins at each voxel. While it is now possible to acquire terabytes of spatial and protein data per cubic centimeter, progress is limited by the ability to convert this data into maps of biological processes that can be quantified and compared. The biomedical community needs scalable software to analyze and visualize whole-organ cellular-level multi-channel images.
Tissue microenvironments contain sparse and space-filling structures such as (1)heterogeneous cellular distributions, (2) interconnected networks of neurons and microvessels, and (3) long muscle, axonal, and connective tissue tracts. This unique complexity requires new analysis and visualization capacity that existing tools cannot provide. Current software relies on isosurfaces and volume rendering, which are insufficient for the biomedical community's current goals of (1) creating searchable organ-scale sub-cellular images, integrating both structure and molecular signatures, and (2)profiling differences between large tissue samples. Meanwhile, researchers continue to acquire terabyte-scale tissue images while lacking appropriate tools for analysis.
Fortunately, specialized algorithms have been developed to address some of these specific challenges in other fields, such as high-performance computing, astrophysics, and computer graphics. A unified framework integrating these algorithms requires a substantial cross-disciplinary effort to assemble, adapt, and optimize domain-specific components.
This Shonan meeting will develop a roadmap for integrating and optimizing algorithms from multiple disciplines into a unifying framework for analyzing, visualizing, and profiling complete whole-organ tissue images. Meeting organizers are leading experts in (1) parallel algorithm development and deployment, (2) visualization of massive data sets, (3)segmentation and analysis of complex structures, and (4) three-dimensional high-throughput microscopy. Participants will include researchers, software developers, and stakeholders in this field - including imaging, clinical, and biomedical specialists.
This meeting will produce practical guidance for a software framework enabling petabyte-scale data composition, analysis, and visualization. This guidance will enable a global community of researchers, developers, and domain experts to independently contribute compatible solutions to address this complex and growing need.
Seminar Aims and Topics
A new field in informatics is necessary to cope with the emergence of massive data sets describing whole-organ tissue at the microscopic and nanoscopic scale. Our goal is to formalize this new field, which exists at the intersection of parallel programming, data structures, data mining, and visualization.
We have identified four topics that will guide a productive discussion:
- Data Structures that facilitate storage, processing, and visualization of massive 3D data composed of sparse volumetric distributions and complex surfaces. These data structures will encode various features from graph theory, 3D geometry, topology, and sparse volumetric representations. Proposed data structures must also facilitate data storage (long-term/cloud), parallel processing (multi-core/GPU), and visualization (rasterization/ray-tracing) for data sizes spanning terabytes to petabytes.
-
Scalable Algorithms that leverage the properties of tissue microstructure, including sparsity and connectivity. Scalable algorithms must take advantage of parallel architectures, such as multi-core processors and GPUs. They must also account for compression and sparse encoding to enable practical data storage, processing, and visualization.
-
Features and Metrics that characterize complex structures to facilitate data mining, quantification, and comparison. Tissue microstructure is inherently complex, containing interconnected networks and protein distributions that are challenging to quantify with existing metrics. Domain experts play a key role in identifying features of interest for individual phenotypes, such as measuring network connectivity. However, converting these intuitive - and often subjective - features into quantitative measurements is challenging, particularly at scale. It is therefore critical to provide a framework for identifying and quantifying complex features and their relationships in tissue microstructure data.
-
Visualization Methods conveying both the structure and subtle features of complex tissue microstructure. We expect effective visualization methods to operate on two levels: (a) Direct representation of complex structures, such as microvasculature, neurons, and protein distributions, which will require efficient rendering and ray-tracing methods to cope with massive data. (b) Approaches that convey derived features, such as quantitative variations in microvascular, neuronal, and cellular structure, orientation, and density.
Expected Outcomes
This project will provide the transformational ability to construct three-dimensional searchable models from petabyte-scale images that integrate explicit structures and implicit molecular distributions, enabling tissue analytics at unprecedented scales. Whole organ cellular level tissue atlases will include: (1) sparsity-exploiting data structures that integrate explicit three-dimensional structures and implicit molecular distributions, (2)massively parallel algorithms integrating recent advances in deep neural networks and perceptual grouping, (3) analytics-guided selective visualization methods enabling efficient proofreading and interpretation. This meeting will produce a comprehensive framework for building browsable morpho-molecular cellular-level and whole-organ tissue atlases.
The roadmap for developing of this framework will be proposed and the major challenges identified by stakeholders and potential developers. A working group will drive the continuous discussion of this new field after the meeting. Attendees will produce and publish a report outlining the results of the meeting. This will include a long-term plan to secure external funding, host additional workshops, and publish a manuscript, most likely a comprehensive book, to guide the development and expansion of this new field.